Methods
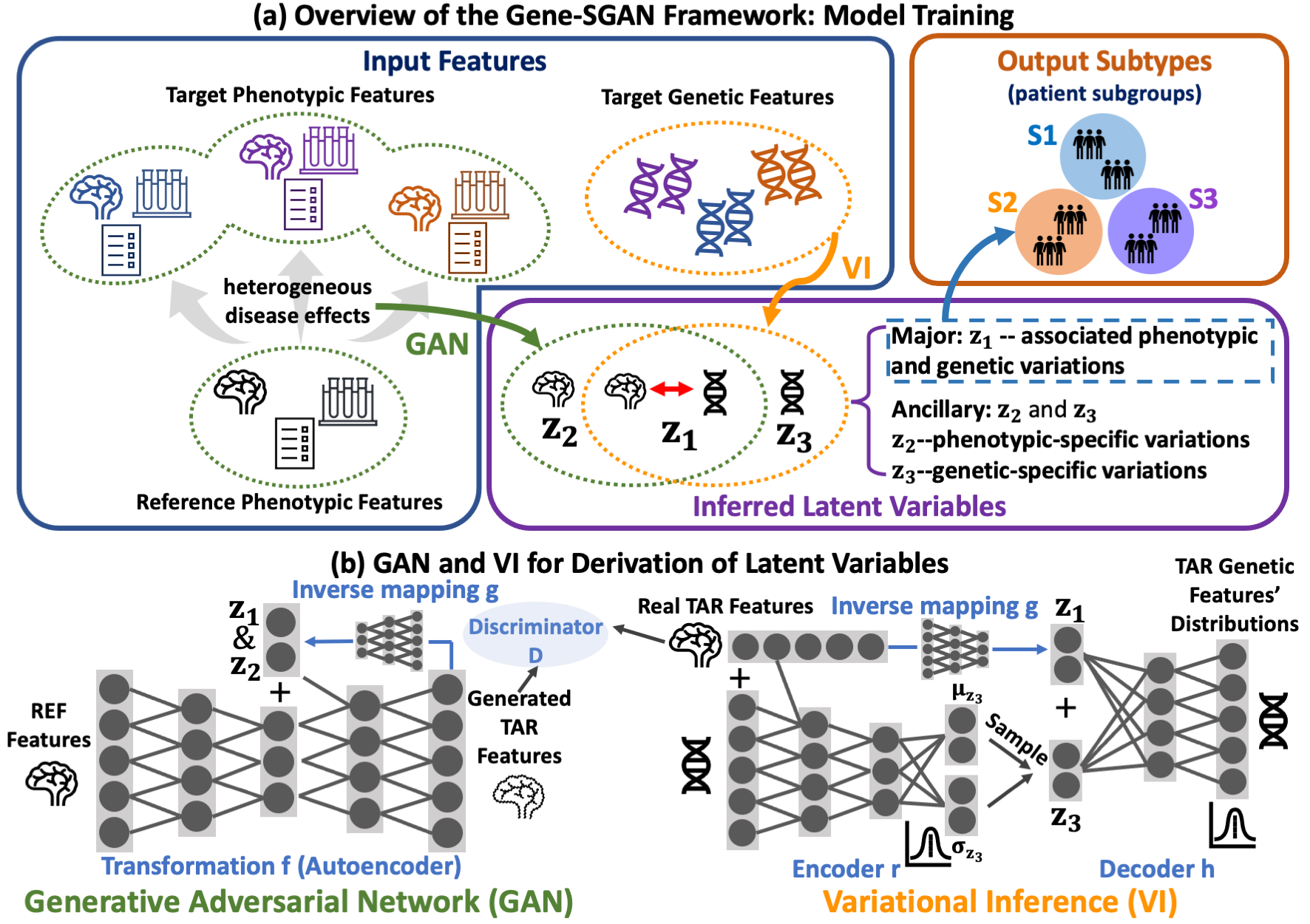
Gene-SGAN: Gene-guided weakly-supervised clustering via GAN
Gene-SGAN is a multi-view weakly-supervised clustering method for disentangling disease heterogeneity. By jointly considering brain phenotypic and genetic data, Gene-SGAN identify disease subtypes with associated phenotypic and genetic signatures. The methodological advances of Gene-SGAN are two-fold. First, through learning one-to-many mappings from phenotypic measures of a reference population (e.g., healthy controls) to a target population (e.g., patient cohort), GeneSGAN captures the diverse patterns of brain changes related to disease, while reducing confounders from disease-unrelated variations. Second, information disentanglement is achieved through latent space in Gene-SGAN, which separately encode phenotypic subtypes associated with genetic factors through the variables z1, while capturing unlinked phenotypic and genetic variations via two ancillary sets of variables z2 and z3, respectively.
Publication SoftwareApplications
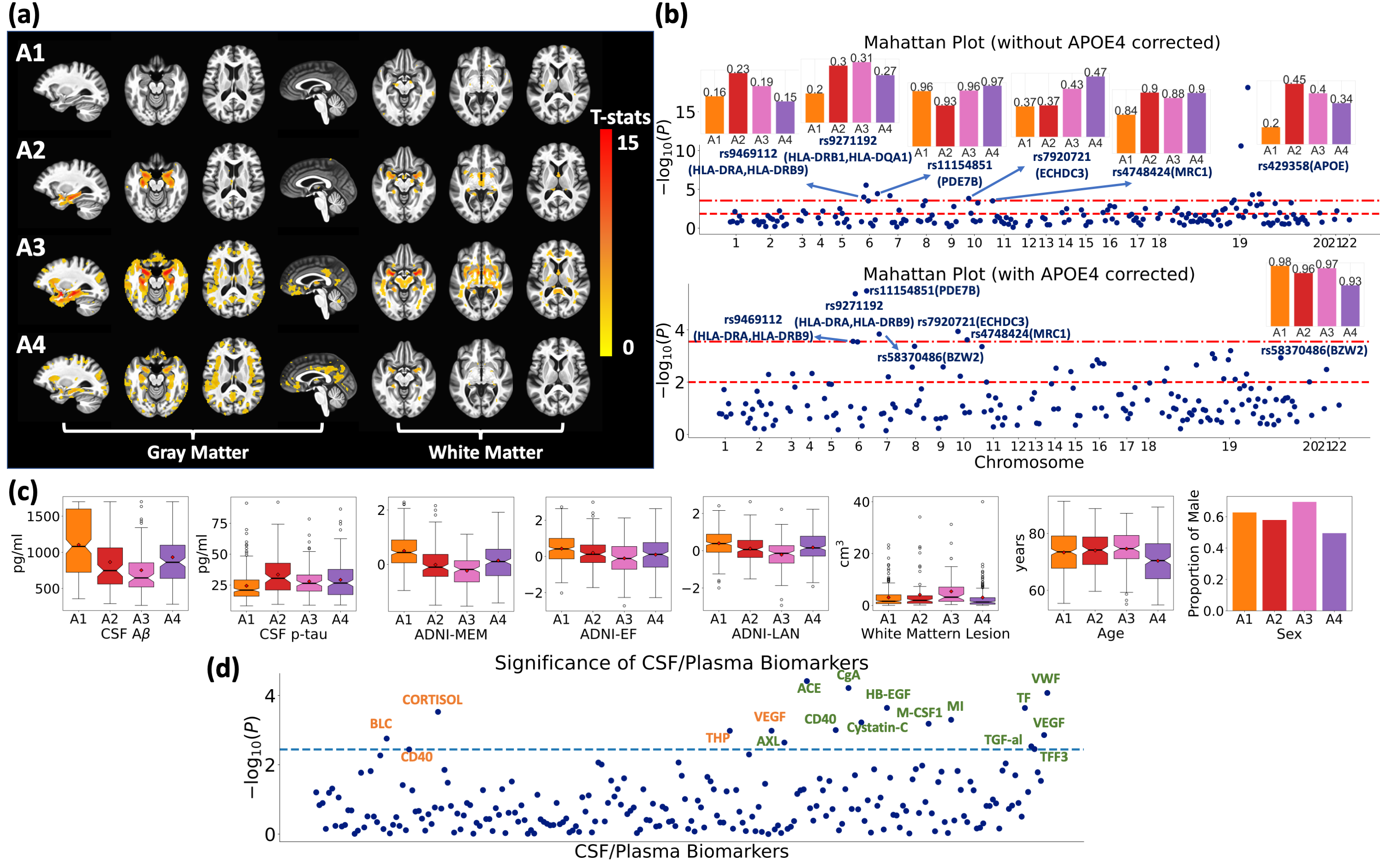
Gene-SGAN identifies four subtypes of brain changes related to AD
Gene-SGAN identified four AD-related subtypes with distinct brain atrophy patterns, demographics, and cognitive performances. Notably, the four subtypes reveal significant differences in seven known AD-related genetic variants. The identified subtype-associated SNPs not only resemble previous findings, but also suggest genetic protective effects contributing to observed resilience, potential inflammatory mechanisms underlying subtypes, and the effect of SNPs on atypical atrophy patterns of AD. These four genetically-associated subtypes also possess significant differences in a large set of plasma/CSF biomarkers, which are related to distinct biological mechanisms contributing to the heterogeneity of AD. For further details, refer to the work of Yang et al.
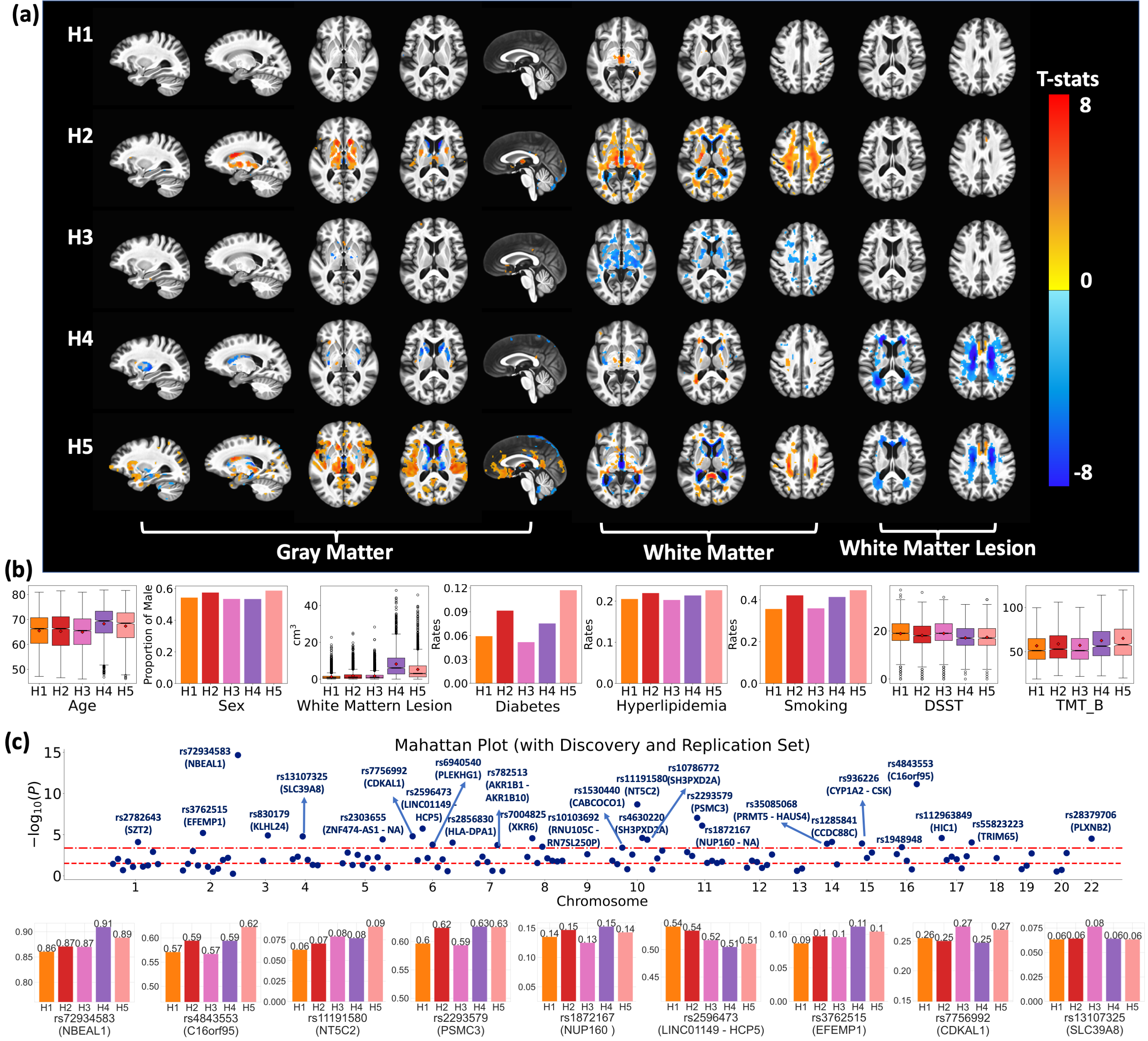
Gene-SGAN identifies four subtypes of brain changes related to AD
Gene-SGAN identified five clinically distinct hypertension-related subtypes, which reflect the remarkable heterogeneity of the effects of hypertension on brain structure. The five subtypes resemble the previously reported associations among blood pressure, comorbidities, and neuroanatomical changes, potentially elucidating heterogeneous effects from various underlying hypertension-related or hypertension-inducing pathological processes. The subtype-associated SNPs were previously associated with WMH volumes and the comorbidities of hypertension (e.g., diabetes and obesity), consistent with the subtypes' characteristics and neuroanatomical patterns. For further details, refer to the work of Yang et al.